InterPred: a web server to identify
and model protein-protein interactions
About InterPred
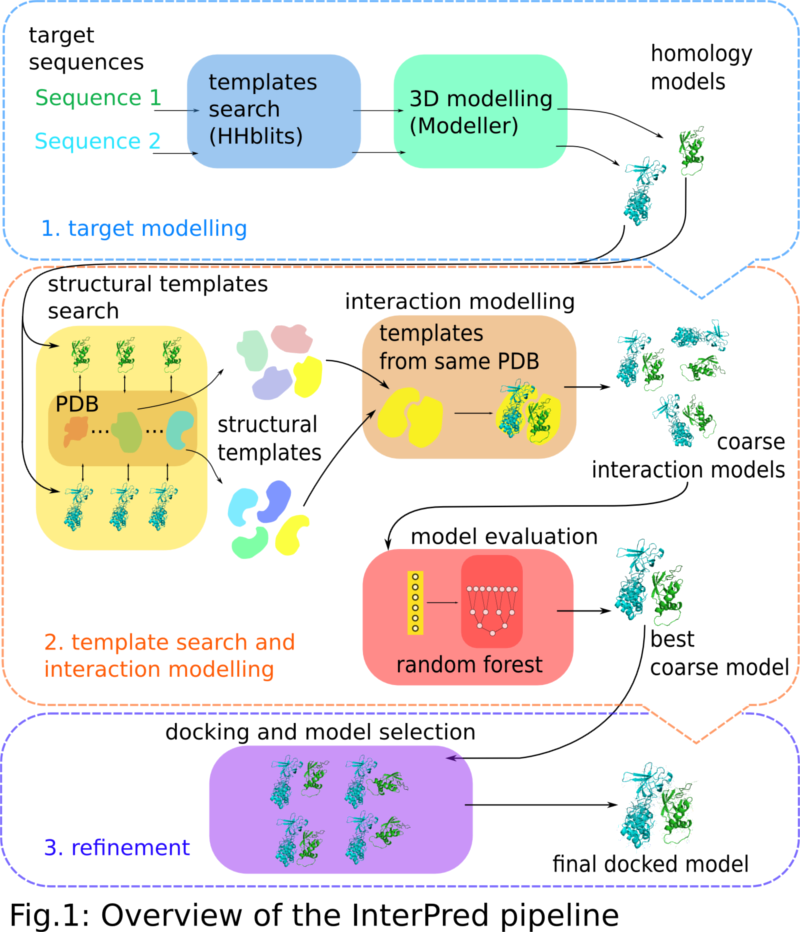 What is InterPred
What is InterPred
InterPred is a web server based on this study: Mirabello and Wallner, 2017. The main goal is to decide if two proteins interact and if so, predict the molecular details of the interaction by building a refined interaction model (i.e. a dimeric molecule from two monomeric molecules).
In short, the InterPred pipeline consists of three steps (Fig. 1):
- Target homology modeling;
- Template search including interaction modeling/scoring; and
- Refinement.
In the first (optional) step the user can submit two target protein sequences. These are used to build structural models using homology modeling. This step can be skipped if the user already has models or experimental structures in PDB files.
Next, structural alignments are used to find close and distant structural similarities to the two models in the Protein Data Bank (PDB). Whenever structures that are similar to the two models also form a complex in the PDB, it defines an interaction template for modeling the interaction. An interaction model is then built by superimposing the representative structures to their corresponding structural homologs in the interaction template. Based on the interaction model a set of features is calculated and used as input to a random forest classifier trained to sift out the more promising models that will go through to the final refinement step.
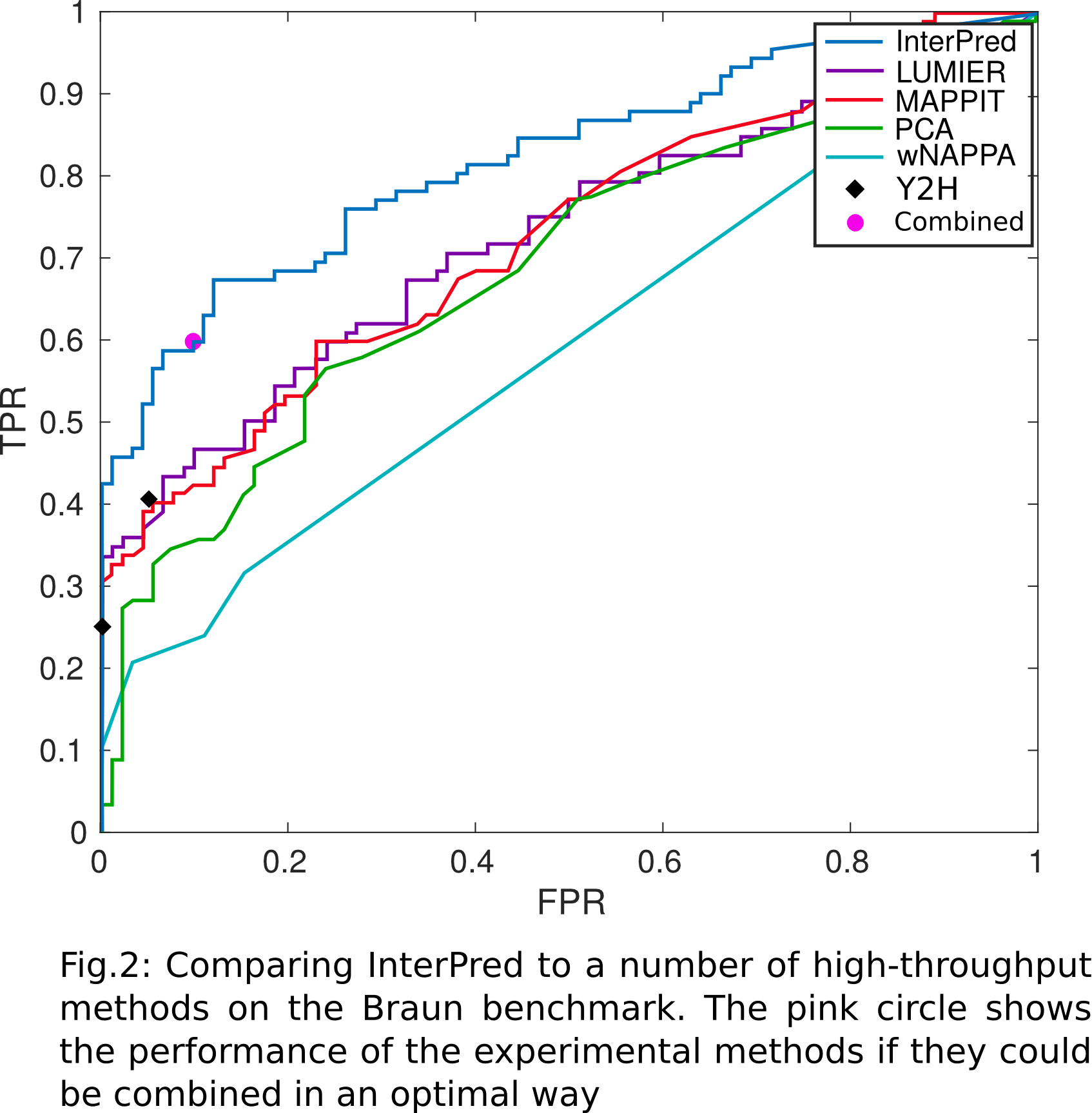 Performance: protein-protein interaction modelling
Performance: protein-protein interaction modelling
InterPred has been tested on a number of independent benchmarks:
It was one of the top three predictors at the latest CASP12/CAPRI37 joint experiment.
It was tested on the Protein-Protein Docking Benchmark and compared favorably with PRISM, another web server for the template-based prediction of protein-protein interactions, and ZDOCK, one of the best ab-initio protein docking methods in literature (see References).
Performance: protein-protein interaction detectionInterPred has also been compared with a number of experimental methods for protein-protein interaction detection. Our results show that it performs better (fewer false positives for any number of true positives) of all of them taken singularly, and at the same lever of an ideal predictor combining all said methods in an optimal way (Fig. 2).
How to use the InterPred web server
Job SubmissionThe user submits to the service two PDB files (click here to see an example of input PDB files) or two target sequences in fasta format (click here to see an example of input fasta files). This can be done through the submission form.
If the user already has two protein structures in a PDB file, these can be uploaded through drag-n-drop to the "Upload your own PDB files" form. If the PDB structures contain more than one chain, it is possible to select the target chains from each file. Otherwise, InterPred will automatically model the interaction of the first chain in each file.
If the user has only the two protein sequences in fasta format, those can be pasted in the two text boxes in the "Paste your fasta sequences" submission form. Note that you need to input your MODELLER license key in the form to use this functionality. If you don't have a MODELLER license key, you can obtain one here (free of charge for academic users or non-profit istitutions).
"Faster" optionBoth forms allow to select the "faster" interaction modelling option to skip the relaxation step on predicted structures.
By deselecting the checkbox, InterPred will run rosettaRelax on the predictions, which will take longer but might yield more realistic structures.
Job statusIn any case, the user is required to submit a working email address and will receive a notification when a job has been completed. It is also possible to check the status of one job by using the "Check the status of previously submitted jobs" form. Here, the status of a single job can be checked by submitting the job ID. It is also possible to check the status of all the jobs submitted with a given e-mail address. The status of the jobs is automatically updated every hour.
Results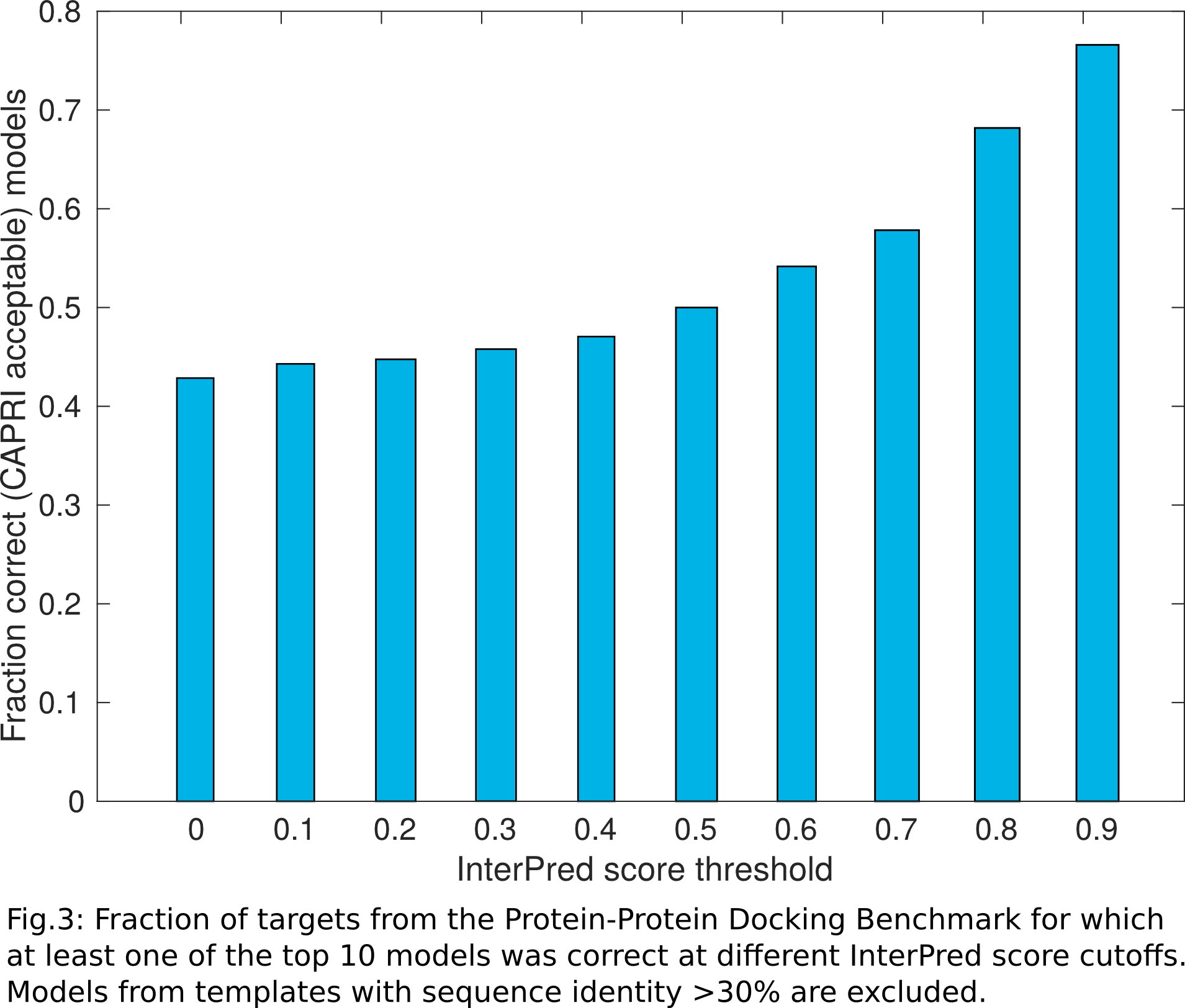
When a job has been completed, the user receives an email containing a link to the results page. Here you can see an example of a results page. The page contains a list of the target monomers - either built from the submitted fasta sequences or provided by the user itself - and a list of refined interaction models. Each model in the list contains information about the templates that were used to build it. Clicking on the corresponding link will launch the download of that model. For each model the InterPred score is also shown. Higher InterPred scores (closer to 1) usually point to higher quality models (Fig.3).
It is also possible to quickly look at each model by clicking on the "View" button. This will open a JSmol applet which can be used to rotate, zoom in/out the molecule, etc. A right click on the applet will open a menu with more useful tools to manipulate the molecule appearance.
If no interaction models are shown, this usually mean that InterPred failed in either modelling the target monomers (if the user submitted fasta sequences) or in finding feasible structural templates for the interaction. If InterPred was run in "fast" mode, it is possible to resubmit the query after deselecting the corresponding checkbox in the submission form. This should increase the chances of predicting the interaction at the cost of a longer waiting time.
References:
Mirabello, Claudio, and Wallner, Björn. "InterPred: A pipeline to identify and model protein–protein interactions."Proteins: Structure, Function, and Bioinformatics 85.6 (2017): 1159-1170.
